Parametrically Constrained Spectrum Analysis¶
This module enables users to perform parametrically constrained spectrum analysis on a chosen experimental data set. The PCSA module fits sedimentation velocity data using linear or sigmoidal constraints to model the relationship between the s and f/f0 of heterogenous macromolecule mixtures. This module is ideal for interacting macromolecules and growing polymer chain systems. Upon completion of an analysis fit, plots available include: model lines; experiment; simulation; overlaid experiment and simulation; residuals; time-invariant noise; radially-invariant noise; 3-d model. Final outputs may include a model and computed noises.
Table of Content:
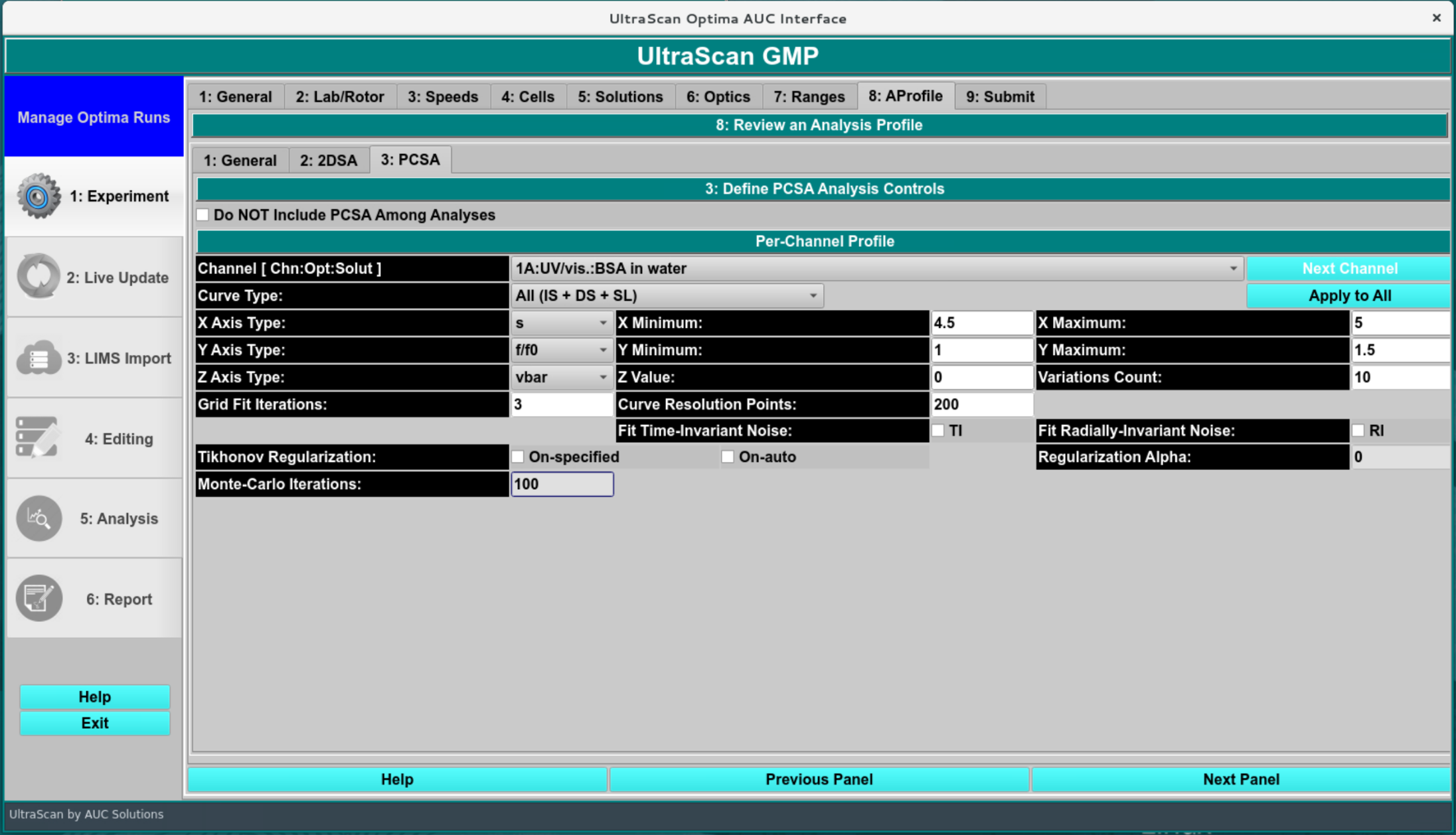
Parametrically Constrained Spectrum Analysis window
Reference:¶
Gorbet G., T. Devlin, B. Hernandez Uribe, A. K. Demeler, Z. Lindsey, S. Ganji, S. Breton, L. Weise-Cross, E.M. Lafer, E.H. Brookes, B. Demeler. A parametrically constrained optimization method for fitting sedimentation velocity experiments. Biophys. J. (2014) vol 106, 1741-50.
Also Available Here